Signal Processing & Computational Biology
Research Focus
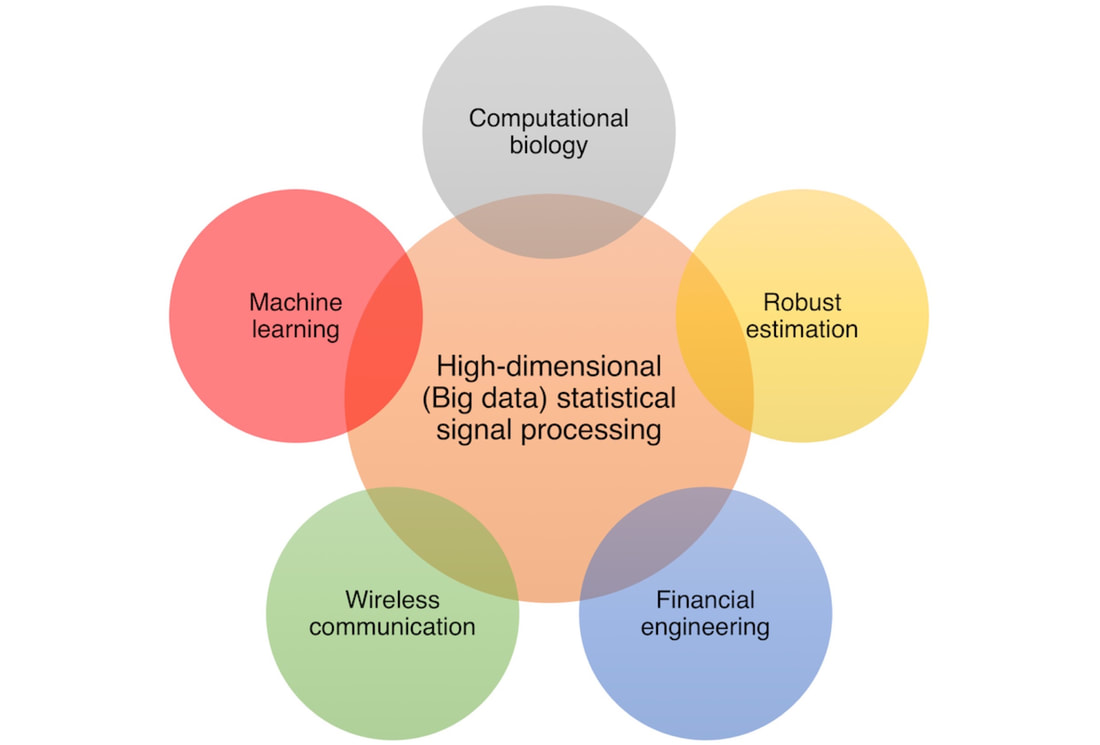
The Signal Processing and Computational Biology Group is an interdisciplinary research group in the Department of Electronic and Computer Engineering at The Hong Kong University of Science and Technology (HKUST). We focus on developing high dimensional statistical and machine learning methods, and applying them to problems in signal processing and computational biology. Our group also works on the theory and applications of random matrices
News
Important group update
Prof. Matthew McKay will be joining University of Melbourne (UoM) as a Professor in the Department of Electrical and Electronic Engineering (EEE), and as an Honorary Professorial Fellow within the Department of Microbiology and Immunology, located within the Doherty Institute. He will retain an Adjunct Professor position at HKUST. The SPCB group will continue to operate at HKUST under Prof. Ahmed Abdul Quadeer. Prof. McKay will continue to work closely with the group at HKUST, while developing a new team at UoM. Research Assistant Professor (RAP) and postdoctoral fellow (PDF) positions are available (see below).
Research Assistant Professor (RAP) and postdoctoral fellow (PDF) positions available
September 2022
Our first monkeypox virus related research “Vaccinia-Virus-Based Vaccines Are Expected to Elicit Highly Cross-Reactive Immunity to the 2022 Monkeypox Virus” has been published in Viruses.
Related bioRxiv preprint: “Vaccinia virus vaccination is expected to elicit highly cross-reactive immunity to the 2022 monkeypox virus“.
Selected media coverage: Medical News Today, South China Morning Post, HKUST/UniMelb/Doherty Institute joint release.
Published paper
Preprint
January 2022
Our COVID-19 research “SARS-CoV-2 T cell responses elicited by COVID-19 vaccines or infection are expected to remain robust against Omicron” has been published in Viruses.
Cover article of Viruses (January 2022 issue).
Related bioRxiv preprint: “SARS-CoV-2 T cell responses are expected to remain robust against Omicron“.
Selected media coverage: The Guardian, The Sun, South China Morning Post, China Daily, Medical News Today, UST/UniMelb joint release.

Published paper
Preprint
December 2021
Hang’s paper “Evolutionary modeling reveals enhanced mutational flexibility of HCV subtype 1b compared with 1a” has been published in iScience.
October 2021
Our research on COVID-19 was highlighted at Advance.org.
Neelkanth’s paper “MIMO Terahertz Quantum Key Distribution” has been published in IEEE Communications Letters.
September 2021
Matthew contributed to the 2021 World Laureates Forum as a young scientist. He presented in a session entitled: “Biology, Medicine and Clinical Sciences”. He also served as a moderator of this session.
July 2021
Upcoming talks:
- Plenary talk at The 11th IEEE International Conference on CYBER Technology in Automation, Control, and Intelligent Systems
(IEEE-CYBER 2021), Jiaxing, China, July 27-21, 2021.
Title: Computational Approaches for Guiding Rational Vaccine Design: Case Studies of HIV, HCV and COVID-19
Speaker: Matthew R. McKay - Talk at Structured Random Matrices in Down Under: New Developments and Applications, Creswick, Victoria, Australia, July 26-August 7, 2021.
Title: Guiding rational vaccine design with random matrix theory
Speaker: Matthew R. McKay
A Theme-Based project “Virological, Immunological and Epidemiological Characterization of COVID-19” on COVID-19 involving our group has been funded by the Hong Kong Research Grants Council. The awarded funding is 60 million HKD.
Matthew is a Co-PI on this project, together with 8 Co-PIs from Hong Kong University (HKU), and 1 Co-PI from the Chinese University of Hong Kong (CUHK). The Project Coordinator is Prof. Leo Poon (HKU).
Related media article: SCMP
Congratulations to Ahmed Abdul Quadeer for his first funded Hong Kong Research Grant Council’s General Research Fund (GRF) project as a principal investigator! Ahmed has been funded 838K HKD and the project is entitled “Identifying targets for a cross-coronavirus vaccine using intelligent computational techniques”. Further details here.
Matthew will be joining 2021 World Laureates Forum as a young scientist.
June 2021
Congratulations to Syed Faraz Ahmed for defending his PhD! Faraz’s thesis was entitled “Genetic Sequence Analysis to Inform Design of Universal Vaccines against Infectious Diseases“. Faraz is going to continue his research as a post-doctoral fellow in our group.

May 2021
Ahmed and Faraz’s paper “Landscape of epitopes targeted by T cells in 852 individuals recovered from COVID-19: Meta-analysis, immunoprevalence, and web platform” has been published in Cell Reports Medicine. As a part of this work, we have developed a web-dashboard that provides an up-to-date information of the emerging data of SARS-CoV-2 T cell epitopes targeted in convalescent patients.
Selected media coverage: MedScape, Reuters; The Medical News; The Tyee;
Related bioRxiv preprint: “Epitopes targeted by T cells in convalescent COVID-19 patients“.
Neelkanth’s paper “Large Intelligent Surfaces with Channel Estimation Overhead: Achievable Rate and Optimal Configuration” has been published in IEEE Wireless Communications Letters.
April 2021
Nicolas’s paper “Large-Dimensional Characterization of Robust Linear Discriminant Analysis” has been accepted for publication in IEEE Transactions on Signal Processing.
Congratulations to Faraz who was shortlisted as a finalist for the SENG PhD Research Excellence Award.
March 2021
Congratulations to Syed Muhammad Umer Abdullah for completing his MPhil! Umer’s thesis was entitled “Methods for Inferring Selection from Short-read Time-series Genomic Data“. Umer will be joining Prof. Kelvin Kai Wing To’s lab at The University of Hong Kong to work on COVID-19 research.
Neelkanth’s paper “Channel Estimation for Reconfigurable Intelligent Surface Aided MISO Communications: From LMMSE to Deep Learning Solutions” has been published in IEEE Open Journal of the Communications Society.

February 2021
Preprint of Hang’s Hepatitis C virus related work “Evolutionary modelling of HCV subtypes provides rationale for their different disease outcomes” has been posted on bioRxiv.
January 2021
Phoom’s paper “Detection of Diabetic Retinopathy from Ultra-Wide Field Scanning Laser Ophthalmoscope Images: A Multi-Center Deep-Learning Analysis” has been published in Ophthalmology Retina. This is a joint work with Fangyao Tang, An Ran Ran, and Carol Y. Cheung (The Chinese University of Hong Kong) and collaborators from Hong Kong Eye Hospital, Sankara Nethralaya and Giridhar Eye Institute (India), Moorfields Eye Hospital (UK), Tel Aviv Medical Center (Israel), and University of Buenos Aires (Argentina).
Our review paper “In silico T cell epitope identification for SARS-CoV-2: Progress and perspectives” has been published in Advanced Drug Delivery Reviews.
Matthew was elevated to IEEE Fellow “for contributions to random matrix theory in statistical signal processing”.
Congratulations to Neelkanth for being selected as one of the three winners of the video contest at the 2021 Global Young Scientists Summit. The summit included recipients of the Nobel Prize, Fields Medal, Millennium Technology Prize, and Turing Award. Neelkanth’s submission “Signal Processing for 6G: From Classical to Quantum Communications” was selected from around 110 entries, all from young scientists who had been selected for the forum.
December 2020
Saqib’s paper “MPL resolves genetic linkage in fitness inference from complex evolutionary histories“ has been published in Nature Biotechnology. This is a joint work with Raymond Louie (University of New South Wales) and John Barton (University of California-Riverside).
Media coverage: UCR Release, Eurekalert.
Neelkanth’s paper “RIS-Assisted MISO Communication: Optimal Beamformers and Performance Analysis” has been published in IEEE Globecom Workshops.
November 2020
Congratulations to Neel for being selected to participate in the 2021 Global Young Scientists Summit.
October 2020
Matthew contributed to the 2020 World Laureates Forum as a young scientist. He gave a presentation in a session entitled: “Learn from the Pandemic: Public Health and Economy”. The session involved 2 Nobel Laureates: Dr. Harvey Alter (2020 Nobel Prize in Physiology and Medicine), and Dr. Ada Yonath (2009 Nobel Prize in Chemistry).
August 2020
Neelkanth’s paper “A Deep Learning-Based Channel Estimation Approach for MISO Communications with Large Intelligent Surfaces” has been published in IEEE 31st Annual International Symposium on Personal, Indoor and Mobile Radio Communications.
June 2020
We have developed a web-based platform, “COVIDep“, for real-time reporting of vaccine target recommendations for the COVID-19 coronavirus (SARS-CoV-2).
- Identification of SARS-derived B-cell and T-cell epitopes that provide vaccine target recommendations for SARS-CoV-2;
- For T cell epitopes, it reports estimated population coverage using HLA/MHC statistical information;
- Intuitive graphical interface with flexible parameter setting;
- Up-to-date reporting based on latest sequence data available (from GISAID).
An article related to the platform “COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2” has been published in Nature Protocols.
Highlighted on the cover page of Nature Protocols (July issue).
Related bioRxiv preprint: “COVIDep platform for real-time reporting of vaccine target recommendations for SARS-CoV-2: Description and connections with COVID-19 immune responses and preclinical vaccine trials“.
Neelkanth’s paper “Signal Design for Frequency-Phase Keying” has been published in IEEE Transactions on Wireless Communications.

May 2020
Saqib and Ahmed’s paper, “How genetic sequence data can guide vaccine design“, has been published in IEEE Potentials Magazine.
Neelkanth, Faraz, and Awais shared their experience as international PhD students at HKUST in a Study International article.
February – March 2020
Faraz’s paper “Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies” has been published in Viruses on 25th Feb. 2020.
Cover article of Viruses (March 2020 issue).
Coverage in World Economic Forum article (by Prof. Wei Shyy, President of HKUST).
Coverage in the cover story (on COVID-19 vaccines) of New Scientist.
Selected press coverage (more on News page): The Standard, Study International, Science Daily, Pharmaceutical Technologies, UST release.

Congratulations to Muhammad Saqib Sohail for successfully defending his Ph.D.! Saqib’s thesis was entitled “A Stochastic Framework for Inference of Population Genetics Parameters from Time-series Genomic Data”. Saqib is going to continue his research as a post-doctoral fellow in our group.

December 2019
November 2019
- “RocaSec: A standalone GUI-based package for robust co-evolutionary analysis of proteins”. Joint with David Morales-Jimenez (Queens University, Belfast).
- “MPF-BML: A standalone GUI-based package for maximum entropy model inference”. Joint with Ray Louie (Kirby Institute, UNSW) and John Barton (University of California-Riverside).
October 2019
September 2019
August 2019
July 2019
June 2019
May 2019
January – March 2019
Our group’s research was highlighted in:
November 2018
September 2018
August 2018

June 2018

Talk at 2018 Artificial Intelligence (AI) in Healthcare Summit, The Ritz Carlton, International Commerce Center, Hong Kong
Title: Inferring the fitness landscape of HIV by unsupervised learning
Speaker: Raymond Louie

May 2018
Title: Using data science to find weak spots of Hepatitis C virus: Implications for rational vaccine design
Speaker: Matthew R. McKay



February 2018
Local, mainland China, and general media
- South China Morning Post – Breakthrough HKUST-led research on mapping of HIV weak spots paves way for vaccine
- RTHK – Hong Kong Today (Big data analysis helping in development of HIV vaccine)
- World News – HKUST Researchers Discover Fitness Landscape of HIV Envelope Protein That May Help Vaccine Development
- Public Now
- Brinkwire – Big data methods learn the fitness landscape of the HIV Envelope protein
- Yahoo – 推算結果 實驗驗證 科大大數據分析病毒 助研發抗愛滋疫苗
Science channels
- Eurekalert – Big data methods learn the fitness landscape of the HIV envelope protein
- Healthcare Analytics News – Big Data and Machine Learning Take on HIV
- Science Daily – Big data methods learn the fitness landscape of the HIV Envelope protein
- Medical Express – Big data methods applied to the fitness landscape of the HIV envelope protein
January 2018
Videos of HKUST
- HKUST Campus, DJI: 2013, 2015, 2016
- HKUST 25th Anniversary
- HKUST Corporate
- Student Diversity
- Hong Kong Ph.D. Fellowship (HKPFS) Scheme (see also here)
Useful Links
- Impact Story on our lab by HKUST’s Development and Alumni Office, http://giving.ust.hk/en/index.php
- Prof. Weichuan Yu’s Bioinformatics and Computational Biology Lab (a partner lab in ECE), http://bioinformatics.ust.hk/
- ECE Department website, http://www.ece.ust.hk
- HKUST website, http://www.ust.hk/
— Banner photo taken by HKUST alumnus Terence Pang
